Review written by Sarah McFann (CBE, G5)
Language is ever-evolving. With each new generation, language structures such as word pronunciation, usage, and meaning mutate and change as they are passed imperfectly from parent to child. Similarly, bodies have the chance to evolve with every generation. Mutations in the germ line—eggs in females and sperm in males—give rise to the genetic variation that allows form and function to evolve. With each new germline mutation, the nucleotides that make up the genetic code are altered due to imperfect DNA replication. These mutations can code for changes to protein structures or protein amounts, altering the way bodies are constructed as they develop and the forms they take on along the way.
In 1790, German writer Johann Wolfgang von Goethe coined the word morphology to describe a way of studying the forms of living things as they change over embryonic development.1 Goethe hoped this new science would shed light on the laws governing organism shape change over time. Now, over two centuries later, we are still trying to understand how organisms change their shapes as they develop, but with additional tools like advanced microscopes and genetics at our disposal. Princeton faculty member Eric Wieschaus, one of the pioneers in the field of developmental genetics, won the 1995 Nobel Prize in Physiology or Medicine for work in this area. His lab is now revisiting the question of morphogenesis—the process through which developing organisms move through different morphologies to obtain their final shape—in a recent paper published in PLoS Computational Biology.2
The Wieschaus lab’s recent paper led by Tomer Stern, a postdoctoral researcher co-advised by Princeton professor Stanislav Shvartsman, describes a new computational method for identifying cell movement motifs in videos of developing tissues. The study focuses on cell movements that occur during gastrulation, the time in development when an organism transforms from a clump of cells known as a blastula into a gastrula composed of different layers of patterned tissue. In gastrulating fruit fly embryos, the model system used in the study, cells that will go on to form the gut invaginate at the front and back of the embryo while cells that will go on to form muscle invaginate from below. Meanwhile, a tissue sheet called the germband sweeps along the embryo surface horizontally, doubling in length as its width is halved. Gastrulation is the first time in fruit fly development where cells have the chance to move in response to genes turned on during the first three hours of embryo growth and thus serves as an excellent model system for studying how tissues change shape in response to gene activation. Stern et al. were interested in better understanding how this dramatic morphological feat is achieved.
Influential work from the Wieschaus lab
Previous work from the Wieschaus lab and others has revealed that the driving force behind germband extension is cell intercalation, a process by which cells change their positions by weakening connections with some neighbors while strengthening connections with others.3 Two of the most common cell rearrangements in germband extension are T1 transitions and rosettes. In a T1 transition, a group of four cells begins in a configuration where the left and right cells touch each other, sharing a vertical boundary. The transition begins when the top and bottom cells start to crowd in from above and below. When the two invading cells meet, they form a new, horizontal boundary, completing the transition. The exchange of vertical boundaries for horizontal ones allows the germ layer to extend horizontally while shrinking vertically. Rosettes are formed in much the same way, except these rearrangements involve five or six cells rather than four.
Back in 1994, Kenneth Irvine, a post-doctoral fellow of Eric Wieschaus, first attempted to pin down the cell rearrangement motifs involved in germband extension.4 Irvine and Wieschaus hooked a video monitor to a simple epifluorescence microscope, taking photographs of the monitor at intervals to catch germband extension in action. They then tracked individual cells and their connections to cell neighbors by repeatedly replaying the video and marking the same cells in different photographs by hand. In this way they were able to identify cell rearrangements. What could not be achieved, however, was a comprehensive list of every rearrangement that occurred in the tissue—it would have been impossible to track every cell and its connectivity to other cells by hand. Such detailed information is necessary, however, to truly understand the processes required to achieve germband extension.
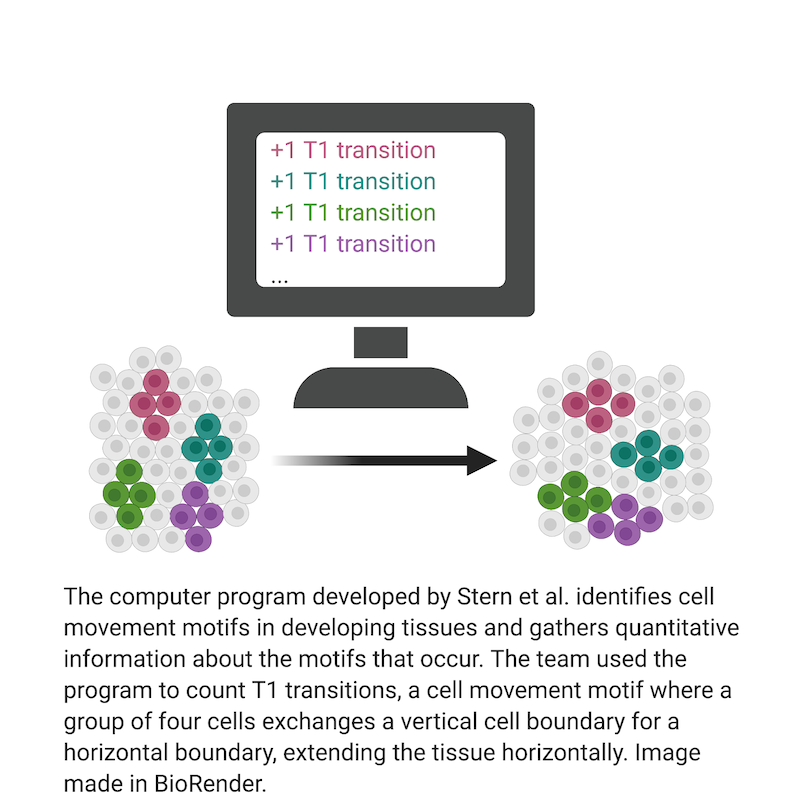
Computational advances
Computational researchers have long since solved the problem of identifying cells in a tissue and automatically capturing features like cell shape and number of neighbors.5 The challenge that remains is to identify cell movement motifs like T1 transitions and rosettes automatically. Stern et al. approached the problem of automatic motif identification similar to the way the program BLAST handles motif matching in genetic sequences.6 The program by Stern et al. translates the cell movement motif the user is searching for into a time series of features. There are many choices of features, like a cell’s roundness or the distance between cell centers, and multiple features can be combined together to search for more complex motifs. Converting a video into a time series of features produces a structure similar to the string of As, Ts, Cs, and Gs that compose genetic sequences. While each position in a genetic sequence represents the nucleotide at a location on a DNA strand, positions in the time series represent the numerical value of a feature at a time point. A method called time warping allows for inexact matches in time, meaning that cell movements that occur faster or slower than the specific motif the user is searching for can still be identified. The ability of this program to find motif matches even in cases of high temporal variability or geometric variability sets it apart from approaches that came before. This study accomplishes what Irvine and Wieschaus, armed with photographs and pens, could only have dreamed of back in 1994.
Describing T1 and rosette transitions
Stern et al. used their program to gain new insights into the precise mechanisms of T1 transitions and rosettes. The team presented their program with three examples of normal germband extension and asked it to identify all T1 transitions and rosettes. With a click of a button, the team was able to count the number of times each motif occurred and visualize the time and place of each cell rearrangement. The program’s T1 count agreed with the count generated by the current gold-standard program for counting T1 transitions, so the team felt confident moving forward and applying their program to more complex analysis. They followed up with a search for a new motif: aborted T1 transitions. While most T1 transitions propel cells in the germ band towards the back of the embryo, a small fraction resolve in the wrong direction and push cells towards the front. By following up with the aborted T1 attempts, the team was able to identify the reason for each failure. They noted that most of the aborted attempts happened in locations where a second morphogenetic event was influencing cell movements, putting germband extension in competition with cell division or an invaginating tissue.
Tomer finds it astounding that this competition between morphogenetic programs doesn’t just result in a big, cellular pileup. He noted, you have all these “cells moving around, intercalating, dividing, delaminating, dying, and changing their shapes to result in a beautifully organized fly larva … [all] at the same time within the same embryo [and] without contradicting the effect of one another.” He explained that even cells sitting next to one another can have different gene activation patterns and thus make completely different movement decisions. “It sometimes seems to me almost like a miracle that development actually works.”
Next, the team followed up on each instance of rosette formation—which occur more rarely than T1 transitions due to the larger number of cells involved. They found that the textbook version of the event, where a group of cells meets up to form a rosette and promptly dissolves, only occurred some of the time. In about 10% of cases “rosette hubs” formed, in which one cell left the rosette as a new cell entered to replace it. This result challenges the traditional idea that all rosettes are transitory events, like T1 transitions. Instead, some locations in the tissue act as long-term hubs for exchanging vertical cell boundaries for horizontal ones. It remains to be determined how and why these hubs form.
Recent advances in microscope technology have made visualizing moving tissues easier than ever before. The advent of lightsheet microscopy has made it possible to watch fruit fly gastrulation from every angle at once. Computational methods like automatic motif identification will prove increasingly necessary to make sense of the terabytes of data generated by such experiments.
Tomer says this is where the project is going next. “Recently we have developed the ability to apply our algorithm to 3D movies of the entire embryo,” he explained. This advancement provides access to the full map of gastrulation cell movements for the first time.
The next challenge is to characterize the frequency and type of motifs that occur in both wild-type and mutant tissues. This would yield a better understanding of how changes to the genome result in changes to the way that tissues perform morphogenesis. Once the occurrences of known motifs are quantified, the focus can shift from automatic quantification to automatic characterization—the search for novel or unknown cell rearrangement motifs.
Organisms all across the animal kingdom use cell rearrangements to achieve their final shape. Identifying how the timing, location, and types of cell rearrangements change as organisms evolve brings us closer to answering the question: what natural processes orchestrate morphogenesis, and what are the rules that determine the sorts of tissue movements that are possible? The recent work described here by Stern et al. represents a notable step forward in the quest to better understand how the language of morphogenesis shifts as genetic codes change.
This article was published in PLOS Computational Biology on August 21, 2020. Please follow this link to view the full version.
References
1Mägdefrau, Karl (1992). Geschichte der Botanik [History of Botany] (2 ed.). Jena: Gustav Fischer Verlag. ISBN 3-437-20489-0.
2Stern, Tomer, Stanislav Y. Shvartsman, and Eric F. Wieschaus. "Template-based mapping of dynamic motifs in tissue morphogenesis." PLoS computational biology 16.8 (2020): e1008049.
3Kong, Deqing, Fred Wolf, and Jörg Großhans. "Forces directing germ-band extension in Drosophila embryos." Mechanisms of development 144 (2017): 11-22.
4Irvine, Kenneth D., and Eric Wieschaus. "Cell intercalation during Drosophila germband extension and its regulation by pair-rule segmentation genes." Development 120.4 (1994): 827-841.
5Khairy, Khaled, and Philipp J. Keller. "Reconstructing embryonic development." genesis 49.7 (2011): 488-513.
6Korf, Ian, Mark Yandell, and Joseph Bedell. Blast. " O'Reilly Media, Inc.", 2003.