Review by Abigail Stanton (MOL, G4)
From the perspective of a cell, every viral infection is the start of a zombie apocalypse. One day you and your neighbor are working together in your quaint body community and then the next… they’ve changed. Your unrecognizable fellow cell becomes single-minded with its new obsession: making viruses. When it has spent all of its energy, it spreads the virus to all of its neighbors, turning them, too, into virus-building zombies.
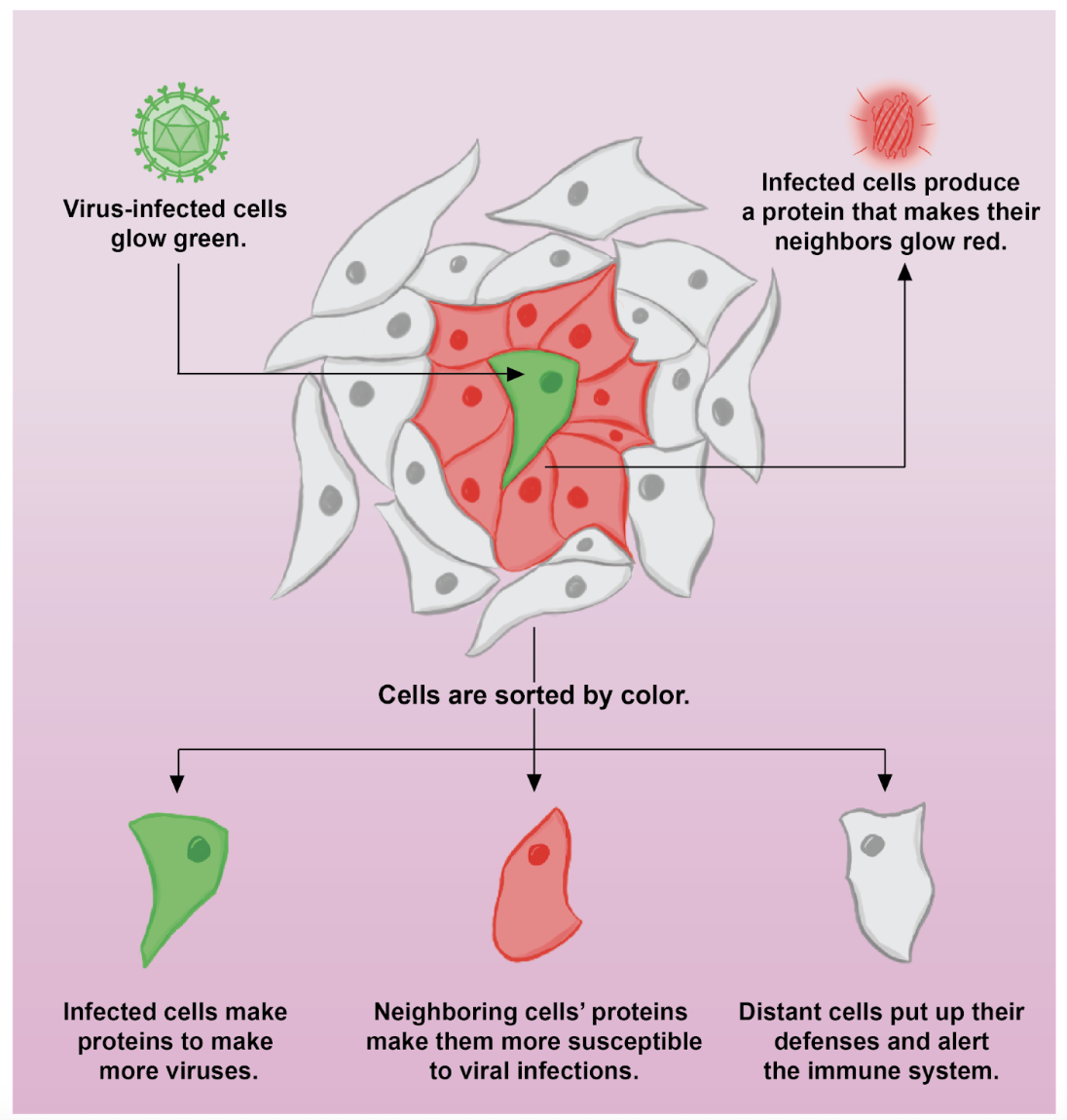
Most viral infection studies only divide cells into two camps: those that have been infected and those that have not. But on the cellular level, there’s a difference between hearing about someone across town being infected and watching it happen to your neighbor firsthand. In their recent publication in Science Advances, members of the Cristea Lab in the Department of Molecular Biology measured how cells respond differently depending on their proximity to an infected cell. To do this, a team led by post-doctoral researcher Bokai Song and graduate student Xinlei Sheng developed a new technique to label cells based on their location relative to a virus-infected cell. They found that cells do, in fact, respond differently when their neighbors get sick–but not necessarily in the ways you might expect.
How to sort cells by where they used to be
When comparing populations of cells, how do we know that they are different? We might be able to use a microscope to see how infected cells dramatically remodel themselves to produce more viruses, but what about the less obvious changes their neighbors may undergo? To get to the heart of what changes in a cell, you need to know what’s going on with its proteins. Proteins are the workhorses of the cell, serving as scaffolding, chemical factories, messengers, and more. Knowing how much of each protein is present can tell you a lot about what the cell is currently prioritizing and what it might be preparing to do next.
The Cristea Lab uses a technique called mass spectrometry (often shortened to “mass spec”) to get a snapshot of all the proteins present in a group of cells. By comparing the proteins in one sample versus another, they can see when protein levels are higher or lower and relate these differences to the various cell functions they are responsible for. However, to gather all the data needed to make these comparisons, thousands of cells need to be removed from where they’re growing and broken down into small components that the instrument can handle. During this process, the cells are all mixed together and any information about where that cell lived in relation to infected cells is lost.
To solve this issue, the researchers took inspiration from a study that labeled cells next to a cancerous tumor.1 Song and Sheng’s team created cells that produce a protein composed of three parts:
- A section that tells the cell to send the protein out of the cell,
- A section borrowed from HIV that allows the protein to enter the neighboring cells, and
- A section that makes the protein glow red.
The resulting protein is exported from the cell and causes any neighboring cells to turn a fluorescent red color. Song, Sheng, and colleagues infected these special cells with their virus of choice, human cytomegalovirus, and mixed them with normal cells that didn’t produce the neighbor-labeling protein. To distinguish between neighbors and the infected cells, they also included a green tag within the virus itself. As a result, in the final mix, infected cells would glow both green and red, neighboring cells would glow red, and cells that grew far away from the infected cell would have no color. Using a technique called fluorescence-activated cell sorting, the researchers could then put the cells in different containers based on their color, allowing them to measure their protein levels separately.
When it comes to viral spread, distance matters
Song and Sheng et al. found distinct differences in the proteins present in the infected, neighboring, and distant cells compared to cells that had never been exposed to the virus. As expected, the infected cells had changes in their protein levels that signaled their transformation into virus-producing factories: increased machinery to copy the virus’ genes, decreased machinery to break down viral proteins, and changes to the cell receptors to make it easier for viruses to enter the cell. This gave the researchers confidence that their technique was sorting cells correctly.
When infected by a virus, cells release chemical signals called cytokines to warn other cells of the danger. Other cells pick up on these signals and bolster their defenses, storing up an arsenal of antiviral proteins to sense and destroy any intruders. The researchers found that the distant cells behaved as expected in response to these warning signs, increasing antiviral proteins and preparing to call in the emergency responders (a.k.a. the adaptive and innate immune systems).
One might think that the neighbors of the zombified infected cells, being exposed to the most warning signals, would be even more likely to raise the alarm and do all they can to protect themselves. However, the data tell a different story: neighbors of infected cells do not activate antiviral defenses as much as distant cells do. In fact, the protein levels in neighboring cells were closer to what was seen in the infected cells. They had an increase in proteins that remodeled the internal compartments of the cell, creating a favorable environment for virus infection. They also had fewer proteins to allow for movement, contact, and communication with their neighbors. As a result of these differences, Song, Sheng, and colleagues found that neighboring cells were more susceptible to infection–not just by the virus used in this study but by other viruses as well. The cells closest to the infected cell, instead of being most on guard, are the most vulnerable.
So why would this be? The researchers noticed that, despite not being infected by the virus, neighboring cells contained some viral proteins. They hypothesized that while the neighboring cells were not making viral proteins themselves, they could still be picking up proteins made by the infected cells. These proteins could overpower the warning signals, causing the cells to make changes to welcome the virus instead of fighting it. Using drugs that blocked the ability of cells to send packages of proteins (extracellular vesicles) to each other, the researchers found that they could significantly decrease the amount of viral proteins in neighboring cells, demonstrating that the virus does, in fact, send proteins in this way.
The findings of this study may seem bleak from the perspective of the cell. While neighboring cells receive warning signs to put up their defenses, viruses have invented a way to send them messages as well, in the form of viral proteins. Just by being close to an infected cell, it’s already too late: the neighbor already started its transformation, lowering its guard and preparing to become a virus-making zombie.
From our more lofty perspective as organisms made of trillions of cells, however, the insights of this work are empowering. As we learn more about how viruses spread, we become more equipped to stop them. The technique developed in this paper can be adapted to study a variety of viruses, allowing researchers to understand the unique strategies viruses use to promote their own spread. Future work could even combine this technique with other rapidly developing technologies to model what infected, neighboring, and distant cells look like when more than one cell type is present or even in a living organism.
The original article discussed here was published in Science Advances on May 10th, 2023. Please follow this link to view the full version.
References
1L. Ombrato, E. Nolan, I. Kurelac, A. Mavousian, V. L. Bridgeman, I. Heinze, P. Chakravarty, S. Horswell, E. Gonzalez-Gualda, G. Matacchione, A. Weston, J. Kirkpatrick, E. Husain, V. Speirs, L. Collinson, A. Ori, J. H. Lee, I. Malanchi, Metastatic-niche labelling reveals parenchymal cells with stem features. Nature 572, 603–608 (2019).
