Review written by Jessi Hennacy (MOL, G5)
Preserving the integrity of DNA is crucial to maintaining a cell’s functions and life cycle. However, DNA is regularly under attack by chemical and physical agents, such as toxins and UV rays from the sun, that can cause breaks in the chemical backbone that holds a strand of DNA together. This DNA damage can lead to dire consequences if left unaddressed, with effects ranging from cell death to uncontrolled cellular proliferation, which leads to cancer. Thankfully, our cells have evolved mechanisms of repairing broken DNA in order to alleviate the risks of accumulating DNA damage.
PARP1, an enzyme that binds to broken DNA, is a key player in DNA repair. After binding to a break in the DNA, PARP1 adds bulky chemical groups called ADP-ribose to many different proteins found in the nucleus, where DNA is housed. This process of adding ADP-ribose is known as ADP-ribosylation. ADP-ribose is added to a specific amino acid within these proteins called serine.
One class of protein targets of the PARP1 enzyme are protein complexes called histones, which act as spools that the string of DNA is wound around. Together, the DNA wrapped around histones form structures called nucleosomes; many nucleosomes packed together make chromatin. Modifications on the individual histone subunits can cause changes to the overall structure of chromatin. Histones are modified by the addition or removal of various chemical groups. These chemical groups can play several roles, including recruitment of other proteins or modulation of how tightly the chromatin is packed. In order to fully understand how chromatin is modified during DNA repair, we need to know what biophysical function these chemical groups play.
Although ADP-ribosylation of histones is associated with DNA repair, it is unknown what specific function this chemical group performs at the biophysical level. Part of the reason for the gap in knowledge is due to a lack of tools to study ADP-ribosylation outside of a cell, or in vitro. In a recent study, postdoctoral scholar Nir Hananya and colleagues of the Muir lab at Princeton address this challenge by designing and executing a set of steps for synthesizing full-length histone proteins with an ADP-ribose group attached at a known serine. Because these synthetic ADP-ribosylated histones are completely chemically defined, they can be compared to otherwise identical histones that lack ADP-ribose in controlled experiments to pinpoint the specific effects that the ADP-ribose group has on chromatin.
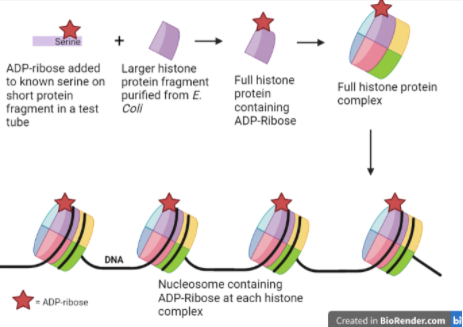
To synthesize ADP-ribosylated histone with a known chemical composition, Hananya et al. first started by synthesizing a very short piece of the histone protein with an ADP-ribose group attached to a specific serine. Synthesizing proteins in a test tube is a time-consuming process and requires a lot of resources, so to obtain a full-length histone protein, the researchers used the bacterium Escherichia coli to produce longer proteins. The bacterium uses its natural protein-generating machinery to create proteins based on the sequence encoded by template DNA that the researchers insert into the bacterium. After purifying the proteins produced by the E. coli, the authors could then chemically ligate, or connect, these large pieces to their original synthesized ADP-ribosylated fragment. This gave them a full-length histone protein with an ADP-ribose group attached to a specific site in the protein.
Hananya et al. then used the ADP-ribosylated histones they made to create artificial chromatin. They started with mononucleosomes, each of which is just a single histone complex with DNA wrapped around it. The composition of these mononucleosomes was examined with a technique called native PAGE (polyacrylamide gel electrophoresis). In native PAGE, a gel composed of a chemical called acrylamide, which serves as a “net” that blocks larger complexes and allows smaller ones to pass, is made in a buffer that protects proteins from falling apart, allowing a complex of multiple proteins to remain intact. An electric current is passed through the gel, and the proteins are pulled through the gel’s matrix at different rates depending on the size of the protein complexes. The larger the protein complex, the further it stays towards the top of the gel, and the smaller the complex, the further it travels to the bottom of the gel.
Native PAGE analysis showed that the ribosylated substrates folded into a complex with the expected eight subunits just as well as the subunits without ADP-ribose did, indicating that they had successfully generated ADP-ribosylated histone complexes. To verify that the ADP-ribosylated mononucleosomes contained ADP-ribose, they treated it with an enzyme that removes the group and showed that this causes their synthesized mononucleosome to behave just like the mononucleosome known to lack ADP-ribose.
Hananya et al. next used their ADP-ribosylated histones to create longer strings of nucleosomes, which could be used to test chromatin properties. These nucleosome arrays were used in an assay to test how compact the chromatin was, and the authors found that the chromatin made with ADP-ribose was less compact. This could make the DNA in the chromatin more accessible to enzymes involved in DNA repair or other processes such as replication.
Finally, Hananya et al. studied the interplay between ADP-ribosylation and other chemical modifications to histones. One example of a post-translational modification that occurs on histones is the addition of a small methyl group to the amino acid lysine, which is performed by enzymes called lysine methyltransferases (KMTs). The authors treated their ADP-ribosylated histones with different KMT enzymes and used a Western blot to assess whether methylation could still occur with the ADP-ribose group attached. This Western blot used an antibody that specifically binds to the methylated sites on the histones, which can then be labeled via binding to a secondary antibody conjugated to a chemical that can be visualized. This experiment revealed that while the ADP-ribose group didn’t completely get rid of methylation, it did decrease methylation at one particular site by 85%. This site for methylation was right next to the serine with the ADP-ribose group. The authors therefore discovered that the inhibition of methylation at that site is likely due to ADP-ribose blocking the enzyme that methylates the lysine (G9a).
With this work, Hananya et al. created a new tool for examining the function of ADP-ribosylation in DNA repair. They were able to synthesize histones containing ADP-ribose at a specific site, demonstrate that ADP-ribose groups loosen chromatin, and provide evidence that these groups may also inhibit methylation of nearby amino acids. This work has important implications in understanding how DNA is repaired by the enzyme PARP1 and lays the groundwork for future studies on how ADP-ribosylation of histones can impact chromatin structure.
Hananya adds: “Since we now have access to a tailor-made ADP-ribosylated chromatin, we plan to expand our investigations regarding the role of chromatin ADP-ribosylation. Specifically, we are trying to discover new protein networks that originate from this unique chromatin modification. This research was not possible without the availability of the advanced instrumentation in the Biophysics Core Facility in the Chemistry Department, and the help of facility manager Venu Vandavasi.”
The original article described here was published in Journal of the American Chemical Society on July 15, 2021. Please follow this link to view the full version.