Review written by Jessi Hennacy (MOL, G4)
All plants use the enzyme Rubisco to capture CO2 during photosynthesis, but Rubisco is hindered by a slow reaction rate and a counter-productive reaction that happens when the enzyme binds to oxygen instead of CO2. Algae, however, have a special organelle called pyrenoid that helps Rubisco capture CO2 more efficiently. Whereas most plants need to express high amounts of Rubisco to capture enough CO2 to grow, the pyrenoid supplies Rubisco with concentrated amounts of CO2 to improve the enzyme’s CO2 capturing activity. If a pyrenoid could be genetically engineered into crops, it could be possible for the plants to capture the same amount of CO2 with less Rubisco, thereby helping them grow with fewer resources. However, this advancement requires understanding the functional roles of proteins involved in building a pyrenoid.
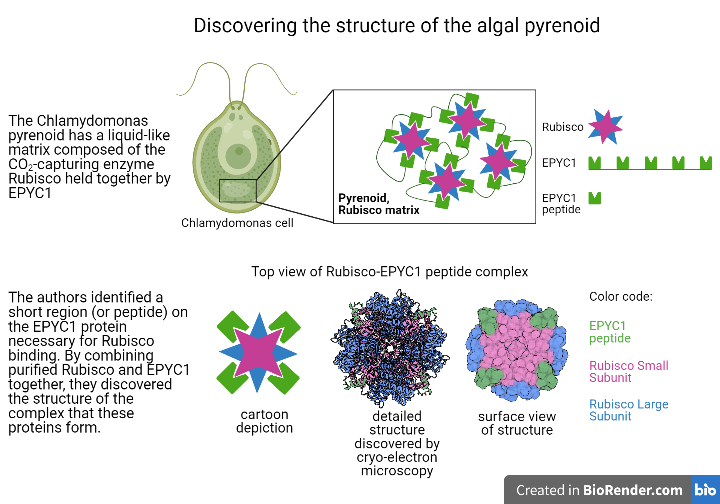
In the algal model organism Chlamydomonas reinhardtii (Chlamydomonas hereafter), pyrenoids are made up of Rubisco enzymes clustered together into a liquid-like region by EPYC1 (Essential PYrenoid Component 1). This region, named the Rubisco matrix, is traversed by membranes thought to deliver concentrated amounts of CO2 to the pyrenoid. EPYC1 is both necessary for a pyrenoid to form in cells1 and sufficient to make liquid-like droplets when added to purified Rubisco in vitro (or in a test tube)2. These in vitro droplets display similar physical properties to the Rubisco matrix found inside Chlamydomonas cells, implying that EPYC1 and Rubisco are all that is needed to form a Rubisco matrix. Understanding exactly how EPYC1 interacts with Rubisco to create the liquid-like matrix region is therefore critical for understanding how the pyrenoid as a whole is assembled, and how this assembly is regulated.
Shan He, a postdoc in the Jonikas lab at Princeton University, led a group that set out to characterize the specific interactions that allow EPYC1 to function as a linker for Rubisco. The liquid-like droplets formed by EPYC1 and Rubisco are unsuitable for structural studies, as the standard structural methods rely on crystallization of individual proteins or protein complexes. However, the authors still needed a way to examine the structure formed when the two proteins are bound. To get around this issue, He et al. first identified the Rubisco-binding site on EPYC1. Because a large portion of EPYC1 is predicted to be intrinsically disordered (flexible, rather than rigid), they hypothesized that the region of the protein that participates in binding would be relatively short, on the order of 18 amino acids long. The team therefore produced short pieces of protein, or peptides, that were each 18 amino acids-long and together spanned the length of the full EPYC1 protein. These EPYC1 peptides were then used to probe for an interaction with Rubisco. They did this using a peptide tiling array; peptides were immobilized on the surface of a grid, and Rubisco purified from Chlamydomonas was incubated with the grid, allowing time for the proteins to bind. Any areas on the grid where Rubisco bound were identified via Western blotting with an antibody that recognizes Rubisco, allowing the team to pinpoint the 18 amino acid region of EPYC1 responsible for binding Rubisco.
Once the EPYC1 peptide that interacts with Rubisco was identified, He et al. used cryo-electron microscopy (EM) to view the complex formed between Rubisco and the EPYC1 peptide. Rubisco is composed of eight identical large and small subunits, and the cryo-EM structure revealed that the EPYC1 peptide binds to each of Rubisco’s small subunits. The sequence of the Rubisco-binding peptide on EPYC1 is repeated five times in the full-length protein, supporting a model where EPYC1 can string Rubisco together in a network within the matrix. Knowing the precise amino acid residues involved in binding also leads to further research into how the binding site can be disrupted by chemical modifications added to EPYC1. Modification of the binding site could be a mechanism through which the cell signals dispersal of the Rubisco matrix, which occurs during cell division, or condensation of Rubisco into the matrix, which occurs during exposure to low CO2.
Having found the potential regions on EPYC1 and Rubisco that mediate binding between the two proteins, He et al. then wanted to test how relevant this information is to the full EPYC1 protein and to forming a Rubisco matrix with the cell. To do this, they purified EPYC1 proteins containing mutations that disrupt their binding sites and added them to purified Rubisco in a test tube. Whereas the normal, or wildtype, EPYC1 is readily able to form droplets with Rubisco, mutant versions that disrupt the Rubisco-binding site could only form droplets at lower salt concentrations. This experiment emphasizes how the binding site they found in the peptide is indeed needed in the full length protein.
They also transformed mutant versions of the Rubisco small subunit. These versions replaced key residues that bind EPYC1 into photosynthesis-deficient Rubisco mutants. The ability to perform photosynthesis at elevated levels of CO2 was recovered, indicating that the mutant Rubisco was functional at fixing CO2. However, these strains containing mutant Rubisco failed to form a pyrenoid or grow efficiently at air levels of CO2, indicating that the EPYC1-binding residues on Rubisco identified in this paper are needed for a pyrenoid to form within the cell.
The structural insights revealed by He et al. advance our understanding of how the pyrenoid is formed through interactions between Rubisco and EPYC1. Understanding the basic biology of the pyrenoid could lead us towards the goal of engineering a functional pyrenoid into crop plants. This addition would improve the efficiency of Rubisco’s CO2-capturing activity, improving the growth of plants and crops.
First-author of the project, Dr. Shan He, said that the equipment and technical support from the Biophysics facility and the Crystallography facility at Princeton were large contributors to this work. Specifically, she acknowledges the great contributions of Phil Jeffrey from the Crystallography facility.
When asked about her future work, Dr. He said she will continue studying “other important components of the pyrenoid which are related with EPYC1 and Rubisco”, as well as examining the regulation of the interaction of EPYC1 and Rubisco.
This original article was published in Nature Plants on November 23, 2020. Please follow this link to view the full version.
References
1Mackinder, L. C., Meyer, M. T., Mettler-Altmann, T., Chen, V. K., Mitchell, M. C., Caspari, O., . . . Jonikas, M. C. (2016). A repeat protein links Rubisco to form the eukaryotic carbon-concentrating organelle. Proceedings of the National Academy of Sciences, 113(21), 5958-5963. doi:10.1073/pnas.1522866113
2Wunder, T., Cheng, S. L., Lai, S., Li, H., & Mueller-Cajar, O. (2018). The phase separation underlying the pyrenoid-based microalgal rubisco supercharger. Nature Communications,9(1). doi:10.1038/s41467-018-07624-w