Review by Tamjeed Azad (COS, G1)
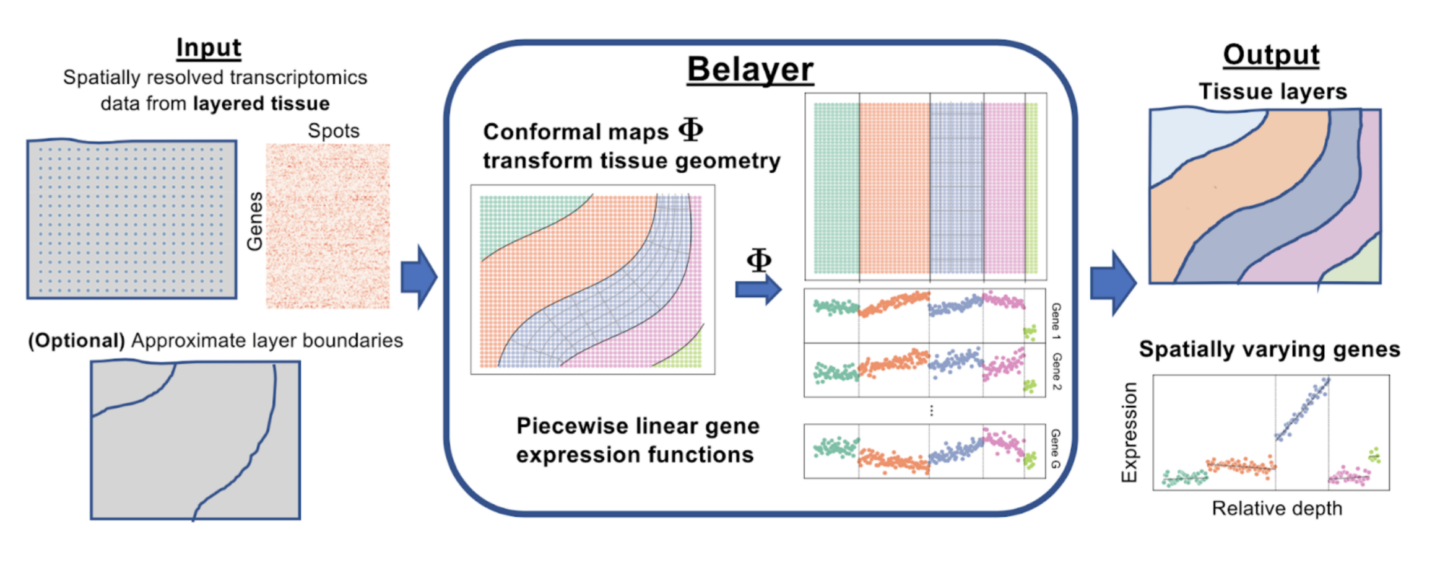
All the marvelous biology we see, from the smallest bugs to the largest trees, is driven by behavior, function, and characteristics of individual cells. The activity of these cells is driven by molecules called proteins, and different cells derive their different characteristics by which specific proteins they use to carry out necessary biological processes. The proteins that are built within cells are determined by which genes of an organism’s DNA are decoded and transcribed into RNA, the blueprint for protein construction. Modern techniques in genetics measure which genes in a tissue are transcribed, in an effort to infer the drivers of such tissues’ biological activity and thus elucidate which functional characteristics are important in a given tissue. Spatially resolved transcriptomics, a subset of these methods, lets biologists and practitioners view which genes are transcribed and to what level (called gene expression) but in a spatial context, making it clear what location within a tissue slice each gene is expressed.
Continue reading “Belayer: Modeling Layered Tissues in Spatial Transcriptomics”