Review written by Abigail Stanton (MOL, G2)
Even in the microscopic world, survival of the fittest can make for relentless, and creative, competition. With a limited amount of resources to go around, some bacteria will play dirty to make sure they get their fair share. Actinoplanes sp. SE50/110, a bacterium that lives in the soil, has developed a strategy to fight off competitors: by producing a specialized sugar called acarbose, it can block proteins responsible for sugar uptake and metabolism in its microbial neighbors. This inhibits the growth of other bacteria, leaving more food for Actinoplanes to enjoy.
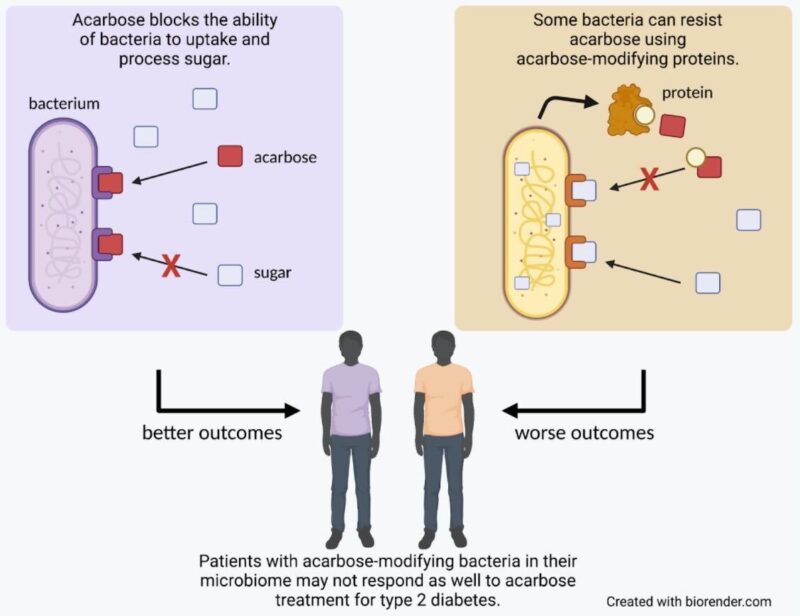
Fortunately for us, while bacteria may use acarbose as a weapon against each other, we can use it to our advantage for therapeutic benefit. When taken orally before meals, acarbose blocks proteins responsible for breaking down complex sugars, reducing the amount of glucose that the body absorbs and aiding in the treatment of type 2 diabetes. However, our bodies also contain a diverse community of microorganisms, our microbiome, which can be susceptible to the effects of acarbose. In a publication out of the Donia lab in the Department of Molecular Biology at Princeton, recently graduated PhD student Jared Balaich and colleagues explore the way acarbose interacts with the human microbiome, identifying a set of genes that allow bacteria to resist its effects. The team’s findings indicate that these genes not only affect how the microbiome responds to acarbose treatment, but may also impact the effectiveness of the treatment for the patients themselves.
What makes a bacterium acarbose-resistant?
To determine why some bacteria are acarbose resistant, the Donia lab first looked to the source. For Actinoplanes’ strategy to be successful, it has to be able to withstand the effects of acarbose while other bacteria cannot. To do this, in addition to creating the “poison” of acarbose, Actinoplanes also makes its own “antidote”: a protein that modifies acarbose so it no longer blocks cell functions. Balaich and colleagues hypothesized that if other bacteria made a protein like this, they would also be able to resist acarbose.
Searching microbiome genomic and metagenomic databases, Balaich and colleagues identified a set of 70 full-length genes found within human microbiomes that have significant sequence similarity to the gene encoding this acarbose-modifying protein found in Actinoplanes. The researchers used these genes to create and purify a few of the best candidate proteins, and found that they were able to modify acarbose in the same way as the Actinoplanes protein. Furthermore, when they took bacteria from the human microbiome that normally struggled to grow in the presence of acarbose and gave them one of these genes, they would instead thrive as if the drug weren’t there.
So why do bacteria in the human microbiome need these proteins?
The team hypothesized that if bacteria in the human microbiome are making the protein “antidote”, they must be in an environment where the acarbose “poison” can be found. They again searched human microbiome genomic data and found a series of genes encoding proteins resembling those needed to synthesize acarbose. Taken together, these findings suggest that, much like with Actinoplanes in the soil, bacteria within the human microbiome may create acarbose-like molecules to compete with each other, leading other microbes to produce acarbose-modifying proteins to protect themselves.
If bacteria in the body can inactivate acarbose, what does that mean for diabetes treatment?
Balaich and colleagues next asked whether the presence of these acarbose-modifying microbes could impact the efficacy of acarbose in treating type 2 diabetes. Making use of data collected by collaborators for another study, they looked at the microbiome sequences of patients taking acarbose. In this sample, roughly half of the patients’ microbiomes contained genes for proteins likely able to modify and inactivate acarbose, while half did not. Intriguingly, patients with acarbose-modifying microbes did not respond as well to acarbose treatment as those without, indicating that the same proteins that microbes use to protect themselves from the effects of acarbose may also interfere with the drug’s ability to block sugar uptake in humans. While this analysis was done using data from only a few patients and will need to be explored further, this finding shines light on a new dimension to consider when weighing treatment options. When studying the effects of a drug, we must think not only of how it might interact with our body but also with the many microbes that call our body home.
When asked what resources made this study possible, Dr. Balaich credited the computational resources and the breadth of expertise available at Princeton. “The collaborative environment here made it really easy to reach out to other scientists with expertise in areas where I had limited knowledge,” says Balaich. He particularly thanks Phil Jeffrey (Macromolecular Crystallography Core Facility) and the lab of Dr. Alexei Korennykh (MOL) for help with protein crystallography, as well as Abhishek Biswas (Research Software Engineer, MOL) for help with computation. “Their contributions really helped drive this study to the next level.”
The original article discussed here was published in Nature on November 24th, 2021. Please follow this link to view the full version.